Site-selective RNA modification by programmable DNA-small molecule conjugates
November 25, 2025Scientists from the National University of Singapore (NUS) have developed an efficient and versatile strategy for site-selective chemical modification of ribonucleic acid (RNA) by employing a DNA-small molecule catalyst that binds selectively to target regions. This research opens new avenues for precise RNA modification, with significant potential for both fundamental research and the development of RNA-based therapeutics.
Site-selective RNA modification is a long-standing goal in chemical biology. However, existing approaches often suffer from RNA sequence bias, complex technical procedures, or limitations in RNA length, which restrict their programmability, rational design, and robustness. A more accessible and broadly applicable method is highly desirable.
The research team, led by Assistant Professor ZHU Ru-Yi from the Department of Chemistry at NUS has developed a site-selective RNA modification strategy that uses a short deoxyribonucleic acid (DNA) guide linked to a small chemical catalyst called DMAP (4-dimethylaminopyridine). The DNA guide pairs with the target RNA sequence through standard Watson-Crick base pairing, positioning DMAP to carry out a precise acyl transfer reaction at the chosen site. This DNA-DMAP molecule can be produced easily via a chemoenzymatic process using an inexpensive, commercially available DNA repair enzyme, bypassing the technical challenges of previous methods.
The findings were published in the scientific journal Angewandte Chemie International Edition.
Once the DNA guide binds with the target RNA region, it positions the DMAP in close proximity to specific nucleotides, allowing a small chemical tag to be attached at that spot. By simply programming the DNA sequence to target a selected RNA region, researchers can achieve high-yield, selective modification with favorable kinetics. This strategy is broadly applicable to various RNA types, including oligonucleotides, ribosomal RNA, and messenger RNA.
The chemical tags used in this study contain an azide group that acts like a built-in “handle” that makes it easy to attach useful additions such as fluorescent dyes. Using this feature, the team labelled mRNA at chosen sites and tracked its movement inside living cells. Importantly, these labels did not significantly perturb mRNA structure or normal cellular processes, underscoring the method’s biocompatibility and utility for nucleic acid research.
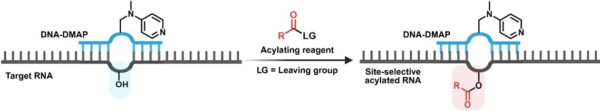
This work introduces programmable DNA-DMAP conjugates that enable site-selective modification of native RNA through proximity-driven catalysis. The approach facilitates targeted acylation of specific 2′-OH groups using PFP esters as acylating agents and is applicable to a broad range of RNA substrates. Moreover, the incorporation of azide-functionalized acyl groups allows subsequent modification via strain-promoted click chemistry, establishing a versatile platform for RNA functionalization. [Credit: Angewandte Chemie International Edition]
Assistant Professor Zhu said, “Our strategy offers a simple, rationally designed approach to achieve site-selective RNA modification without relying on enzymes, ribozymes, or complex solid-phase synthesis. This is the first example of using small-molecule catalysts for this purpose. We believe the simplicity and robustness of our method will make it widely accessible to researchers working with nucleic acids.”
Looking ahead, the team aims to explore new catalysts and reagents to further expand the scope of RNA modifications and apply this approach to endogenous RNA modification in living cells. They also envision that novel RNA structures arising from this strategy will pave the way for efficient RNA-based therapeutics and other advanced scientific applications.
Reference
Kha T.-K.; Zhang T.; Kunchur N.; Zhao Y.; Guo J.; Chen S.; Zhu R.-Y*, “Site-Selective RNA Modification by Programmable DNA-Small Molecule Conjugates” Angewandte Chemie International Edition DOI: 10.1002/anie.202515411 Published: 2025


